Data Curation and Enrichment
We are experts in the curation, enrichment and management of diverse types of real-world health data.
How We Work
Our data engineering, clinical informatics and machine-learning teams have developed a unique data platform that allows us to create research-ready datasets at scale.
By this we mean that health data accessed through our HRA-approved RWD Network is standardised into our common data model, quality controlled and further enriched with other data where possible. The data platform was developed by our Data Engineering and Clinical Informatics teams in close collaboration with our clinical experts and supported through the latest advances in machine learning.
The data platform has been designed to enable the seamless refreshment of datasets to provide the latest disease information from which RWE insights are generated, whilst multiple tools have been developed for deployment across the RWD Network to support efficient, quality-controlled and scalable curation of anonymised data.
Our Advanced Curation Approach
Data Standardisation
Arcturis has established its own proprietary Common Data Model which is compatible with the Observational Medical Outcomes Partnership (OMOP) but is more expansive, incorporating genetic and functional information that is critical to life sciences research and regulatory submission.
Our Common Data Model allows us to work with diverse source data accessed through the RWD Network, harmonising it for the purposes of data quality and ease of analysis.
Data Enrichment
A significant amount of clinical information resides within unstructured clinical notes and reports, impeding direct access to data for analysis.
We have developed ArcTEX (Arcturis Text Enrichment and Extraction), a natural language processing or NLP based model to extract biomarker and other disease specific information from unstructured reports.
ML-Assisted Concept Mapping
We have developed a ML-Assisted Concept Mapping tool which enables us to seamlessly create suggested mappings from any source data to recognised standard concepts, including SNOMED CT and ICD10. These suggestions are then validated by our clinicians, allowing our researchers to benefit from clean, quality concept data.
Regulatory Grade Data Quality Management
The collation of guidance across industry regulators (e.g. FDA, EMA, MHRA), technology appraisers such as NICE and academic research has allowed us to produce a comprehensive Data Quality Framework.
This allows us to report and monitor metrics relating to how plausible and complete data is and whether in conforms to pre-specified data standards.
Free-text PII Redaction
Protecting patient confidentiality is paramount. Our sophisticated redaction tools use state-of-the-art techniques to securely ensure sensitive information has been anonymised, ensuring compliance with privacy laws and ethical standards.
Robust Data Security
Our commitment to data security is integral to our data engineering ethos. We employ cutting-edge techniques to ensure strict compliance standards while facilitating vital research. Our NLP technologies further bolster our capabilities, enabling the extraction of meaningful insights from anonymised unstructured data sources without compromising security.
Arcturis is ISO 27001 and Cyber Essentials Plus certified and is rated as “standards exceeded” for the NHS Data Security and Protection Toolkit.
Introducing ArcTEX
Data enrichment through the use of unstructured clinical reports and other documents.
Our team of researchers have pioneered the development of the NLP model, ArcTEX.
ArcTEX is a sophisticated yet flexible Natural Language Processing (NLP) framework designed to cater to a diverse array of clinical reports.
We have engineered ArcTEX to systematically extract biomarker and disease-specific data from unstructured clinical reports and notes at scale. Notably, ArcTEX stands out for its versatility, and is capable of being easily fine tuned to cater to diverse project needs.
Learn About ArcTEX
Introduction
Numerous studies have demonstrated the vast reservoir of clinical insights hidden within unstructured textual data, such as clinical letters or pathology reports (1). Unfortunately, this highlights the volume of data unavailable for direct analysis. At Arcturis, our team of researchers have pioneered the development of ArcTEX (Arcturis Text Enrichment and Extraction) model. ArcTEXis a flexible Natural Language Processing (NLP) framework, engineered to systematically extract biomarker and disease-specific data from unstructured clinical reports. Notably, ArcTEX stands out for its versatility, and is capable of being easily fine tuned to cater to diverse project needs. Moreover, our model has been meticulously optimised to underpin high-quality real-world evidence (RWE) initiatives, ensuring robust and reliable outcomes across multiple disease areas.
Many leading pharmaceutical companies enhance their clinical development and post-market launch strategies through the integration of real-world data. These can encompass a spectrum of methodologies, ranging from retrospective cohort studies to the optimisation of patient selection criteria or the incorporation of external control arms. However, a significant hurdle lies in the fact that a substantial portion of vital healthcare data required for these initiatives reside within unstructured textual formats, impeding direct accessibility for analysis.
If we consider, for instance, critical biomarker statuses like human epidermal growth factor receptor-2 (HER2) or oestrogen and progesterone receptors (ER or PR). These biomarkers have a profound influence on the treatment trajectory for breast cancer patients. This crucial information often finds itself embedded within unstructured pathology reports, characterized by variations in style and content across different hospital sites and pathologists. This also applies to, for example, nuances regarding ‘response to treatment’ or ‘disease progression’, which further exemplify the breadth of which unstructured data can crucially inform treatment pathways.
Unlocking these insights presents a formidable yet essential challenge in advancing pharmaceutical research and delivering better patient care. To meet the evolving needs of these communities, Arcturis are proud to introduce ArcTEX, a sophisticated NLP model designed to cater to a diverse array of clinical reports. Our innovative solution empowers real-world evidence studies by seamlessly automating the extraction of biomarkers and other disease-specific data at scale. ArcTEX stands as a testament to our commitment to advancing healthcare research through cutting-edge technology, to ensure that the availability of high-quality data is at the forefront of real-world evidence based decision making.
Case Study 1: Extraction of Oncology Biomarkers
Our approach in the creation of our innovative model is based on recent developments in the field of machine learning and natural language processing, utilising transformer-based language models. In contrast to other large language models (LLMs), ArcTEX significantly benefits in not suffering from hallucinations and providing additional confidence scores for each extracted value. The model is also optimised on UK-specific data for a range of clinical markers. Further optimisation is also possible through our iterative optimisation and validation framework, which can provide insights in the robustness of ArcTEX.
As an example, the figure below shows the results of ArcTEX on the detection and extraction of information over a wide range of markers. With an average accuracy of 98.3%, ArcTEX shows the ability to extract multiple markers from free unstructured text reports over both histological and genetic reports. Additionally, ArcTEX is able to extract both numeric-based and text-based values where both are used interchangeably, for example with oestrogen or progesterone receptors amongst others. The versatility of ArcTEX makes it suited to marker extraction from free text reports, extracting results at an accuracy comparable or greater than that of a human annotator (2), at a greatly increased rate of extraction.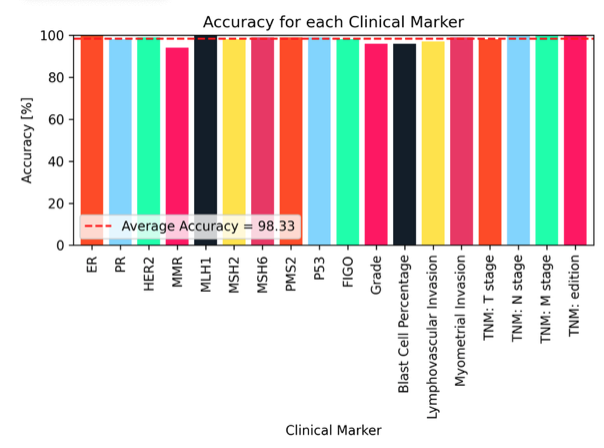
Case Study 2: Model Comparison
In comparison to other available transformer-based language models, such as RoBERTa, BioBERT, or large language models (LLMs) such as Meta’s Llama2 and Llama3 models, ArcTEX has shown superior performance in biomarker extraction. In the figure below, we can see the average overall accuracies of the aforementioned models compared to ArcTEX on the left, with the performance across specific markers displayed on the right. In addition to greater overall performance, ArcTEX also provides insights into the model performance. Unlike with LLMs, for each evaluated free text report, ArcTEX will generate scores based on its confidence that it has identified the correct value. By exploring these confidence scores, reports can be automatically identified which have a risk of having been misclassified. These reports can be flagged for either manual review or excluded from the analysis, resulting in an overall accuracy increase.
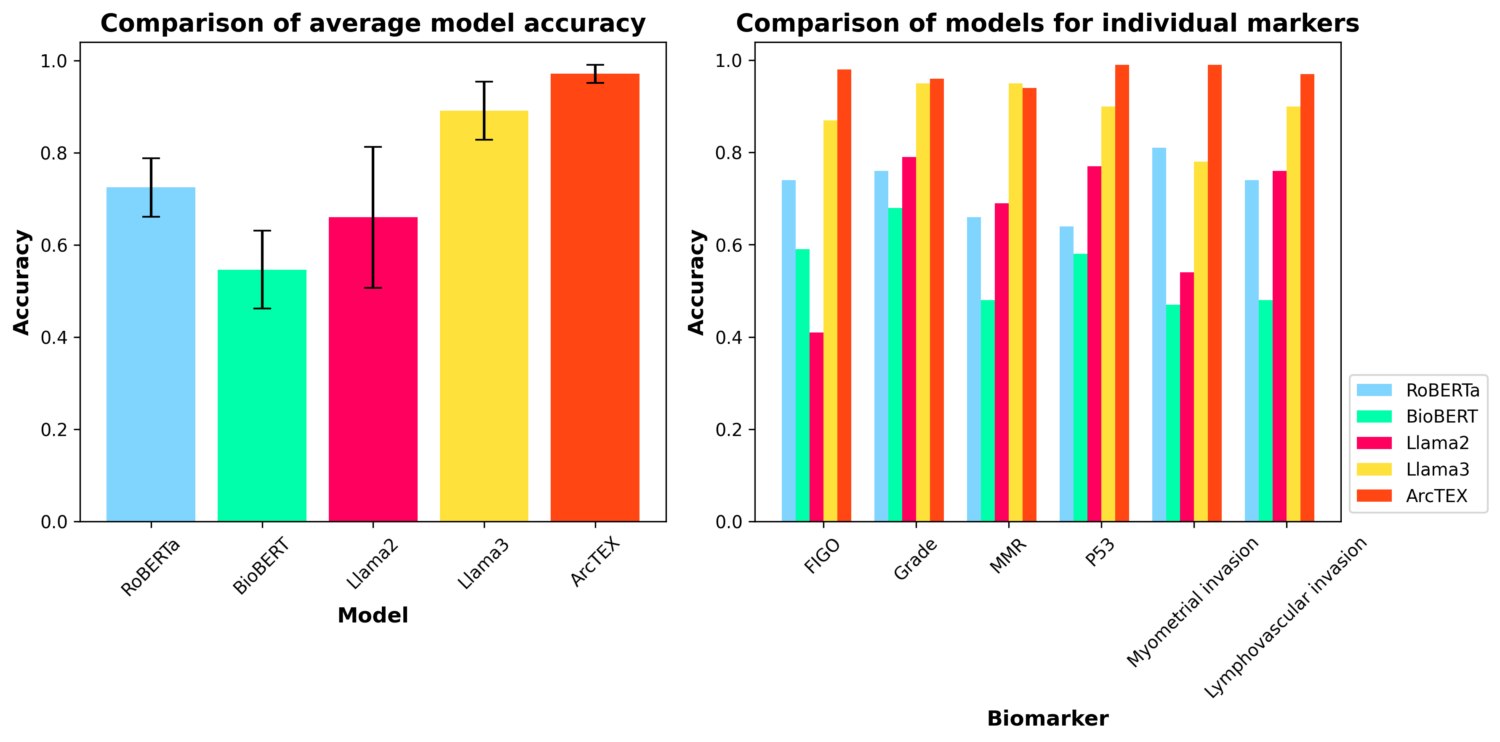
Compared to other LLMs, ArcTEX provides multiple advantages, making it very well placed for a range of RWE studies from a scientific and practical standpoint, such as:
- High accuracy: the model is optimised to extract biomarker and disease-specific data from clinical free text reports which makes the model superior compared to many generic LLMs. The degree of otherwise unavailable granular data that can be extracted and analysed using our model is key to be able to generate useful scientifically robust epidemiological country-specific data to speed up reimbursement and patient access.
- High flexibility: the developed optimisation framework can be used to further optimise ArcTEX on other biomarkers depending on project requirements within hours, which can be highly beneficial from both a cost and efficiency perspective.
- Transparency: ArcTEX provides confidence scores for each evaluated report, allowing quick identification of reports which might require further manual review. Furthermore, the optimisation framework provides insight into the accuracy and variability of the data extraction. This can be used to provide evidence in the robustness of the methodology to support studies.
- Low resource requirements: our approach does not require availability of expensive compute infrastructure like GPU’s. ArcTEX can be executed on a standard computer, which increase the flexibility to deploy our data enrichment tool in different hospital environments.
Case Study 3: Leveraging Clinical Insights for Enhanced NLP Performance
Mismatch Repair (MMR) proteins—MLH1, MSH2, MSH6, and PMS2—play a crucial role in correcting DNA replication errors. Mutations in these proteins can lead to MMR deficiency (dMMR), contributing to genomic instability and impacting treatment decisions in cancers such as endometrial and colorectal cancer. However, identifying patients with dMMR from real-world data is challenging due to the variability in how MMR and its four sub-markers are documented in free-text pathology reports. Reporting practices vary significantly across pathologists, hospitals, and healthcare systems. Some reports provide detailed information on each individual protein, others simply state the overall MMR status as deficient (dMMR) or proficient (pMMR), and some use a combination of these approaches.
Naive methods, such as using basic search techniques like regular expressions, often struggle to effectively capture this variability, resulting in a limited number of patients being correctly identified for analysis.
In a recent analysis of an oncology dataset contained within the Arcturis real-world data. ArcTEX was able to identify 20% more reports with actionable MMR status compared to a naïve implementation by considering both the overall MMR status and all four sub-markers. As shown below, ArcTEX was able to identify 1,606 reports containing MMR information, compared to 1,336 reports using a naive approach. Additionally, ArcTEX can filter out reports where clinical features are mentioned in a non-actionable context, such as “not applicable”, “requested”, or “to follow”.
Of these 1,606 reports, 51.9% contained both the overall MMR status and details about individual sub-markers, 31.4% mentioned only the overall MMR status, and 16.6% listed the individual sub-markers without specifying the overall status. This variability highlights the complexities of MMR reporting, even within a single dataset.
For reports that include both, the overall MMR status and individual sub-markers, ArcTEX further enhances data quality by cross validating this information. As illustrated in the figure above, there is a 97.8% agreement between explicitly stated MMR status and the status inferred from the individual sub-markers when both are available. The remaining 2.2% of discrepancies are flagged for manual review. This process allows for a more efficient and accurate analysis of free text oncology data, ultimately enhancing data reliability and providing deeper insights into oncology research.
Conclusion
In summary, ArcTEX emerges as a pinnacle of innovation in the realm of NLP, offering flexibility and precision in the extraction of clinical features and supplementary disease information from an expansive spectrum of unstructured clinical text. Due to its remarkable capacity to quickly evaluate a vast volume of reports with unparalleled accuracy, as well as integrated robustness matrices and confidence scores, it is an indispensable asset for high-quality RWE generation.
Opportunities for RWE generation exist throughout the entire development cycle, from informing internal strategy (e.g. early-phase target product profile development), all the way to late-stage external control arms, playing a crucial role in regulatory decision making. ArcTEX can be utilised across this entire cycle to unlock the true potential of unstructured data and overcome barriers to analysis, ensuring robust and reliable outcomes for a variety of stakeholders, across multiple disease areas.
References
(1) Hyoun-Joong Kong, Managing Unstructured Big Data in Healthcare System, Healthcare Informatics Research, 2019
(2) Marie-Pier Gauthier et al.: Automating Access to Real-World Evidence JTO Clin Res Rep, 2022
